pCpGfree vitro, A New Family of CpG-free Plasmids
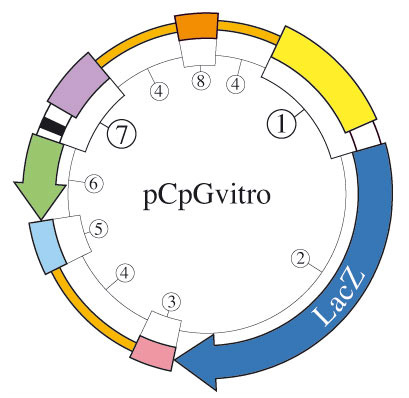
pCpGvitro is a new family of expression vectors completely devoid of CpG dinucleotides that are selectable in mammalian cells.
Similarly to the other pCpG plasmids (i.e. pCpG-lacZ, pCpG-mcs, and pCpG-siRNA), all the elements required for replication and selection of the plasmids in bacteria, and gene expression in mammalian cells have been modified to remove all CpG dinucleotides:
1) CpG-free promoter combining the mouse CMV enhance the human elongation factor 1 core promoter and 5'UTR containing a synthetic intron. This composite promoter yields high and ubiquitous expression of the lacZ gene.
2) CpG-free allele of the lacZ gene. This reporter gene can be easily subcloned and replaced by a gene of interest.
3) and 5) CpG-free versions of the SV40 late and human β-globin polyadenylation signals (polyA). These polyA enable efficient cleavage and polyadenylation reactions resulting in high levels of steady-state mRNA.
4) CpG-free matrix attached regions (MARs) are AT-rich sequences that are able to form barriers between independent expression cassettes.
6) CpG-free antibiotic resistance genes: BlastiR, HygroR, and NeoR. These genes are active both in E. coli and mammalian cells.
7) CpG-free SV40 promoter in tandem with a bacterial promoter located within a synthetic intron. This composite promoter drives the expression of the resistance gene.
8) CpG-free E. coli R6K gamma origin of replication. This origin is activated by the R6K specific initiator protein π.
pCpG vitro plasmids represent innovative tools to study the effects of CpG dinucleotides in a number of applications. DNA vaccination exploits the immunostimulatory character of certain CpG motifs to prime and boost the immune response. However, these immunostimulatory CpG motifs are antagonized by CpG dinucleotides in certain distinct base contexts, termed neutralizing CpG motifs. Both types of CpG motifs are usually present in plasmidic DNAs, and therefore may lead to an unfavorable immune response. pCpGvitro is the ideal tool to overcome this problem, and may be used to study the effects of these two types of CpG motifs by adding them in different configurations to the pCpGvitro backbone.
CpG dinucleotides are key elements in a number of cellular functions associated with chromatin. Many recent findings have altered our vision of chromatin and its role in the regulation of cellular processes such as transcription regulation, DNA replication and repair, cell cycle control, and cell aging. Several large multisubunit complexes, consisting of methyl-CpG binding (MBD) proteins and histone deacetylases, have been implicated in the regulation of chromatin dynamics. These complexes are recruited to methylated CpG dinucleotides by DNA methyl transferases (DNMTs) and induce chromatin remodelling. However the specific roles of these complexes are still to be explored. Due to the absence of CpG dinucleotides within its backbone, pCpGvitro is not the target of DNMTs and thus MBD proteins. Therefore, it provides a useful model to study the other proteins involved in these complexes, in particular the histone deacetylases. It can also be used to analyze the effects of CpG methylation on the regulation and duration of gene expression.



