RNA Interference
RNA interference (RNAi) is one of the most exciting discoveries of the past decade in functional genomics. RNAi is rapidly becoming an important method for analyzing gene functions in eukaryotes and holds promise for the development of therapeutic gene silencing.
RNAi and gene silencing
RNA interference (RNAi) is a post-transcriptional process triggered by the introduction of double-stranded RNA (dsRNA) which leads to gene silencing in a sequence-specific manner. The first evidence that dsRNA could achieve efficient gene silencing through RNAi came from studies on the nematode Caenorhabditis elegans. Further analyses in the fruit fly Drosophila melanogaster have contributed greatly toward understanding the biochemical nature of the RNAi pathway [1].
Long dsRNAs are cleaved by the RNase III family member, Dicer, into 19-23 nucleotides (nt) fragments with 5’ phosphorylated ends and 2-nt unpaired and unphosphorylated 3’ ends.
These small dsRNAs are called small interfering RNAs (siRNAs). Each siRNA duplex is formed by a guide strand and a passenger strand.
The endonuclease Argonaute 2 (Ago 2) catalyzes the unwinding of the siRNA duplex. Once unwound, the guide strand is incorporated into the RNA Interference Specificity Complex (RISC), while the passenger strand is released. RISC uses the guide strand to find the mRNA that has a complementary sequence leading to the endonucleolytic cleavage of the target mRNA [2].
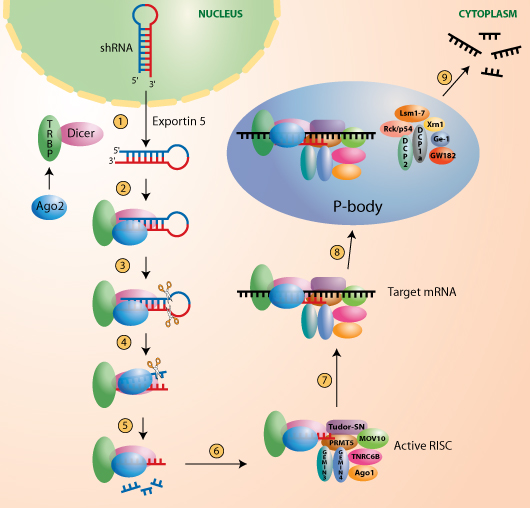
RNAi-mediated gene silencing in mammals using shRNAs
① Plasmid-expressed short hairpin RNA (shRNA) requires the activity of endogenous Exportin 5 for nuclear export [10].
② Ago2 (Argonaute 2) is recruited by TRBP [11], that forms a dimer with Dicer [12], and then receives the shRNA [13-15].
③ The shRNA is cleaved in one step by Dicer generating a 19-23 nt duplex siRNA with 2 nt 3’ overhangs.
④ After identification of the “guide strand” in the siRNA duplex, the “passenger strand” is cleaved by Ago2 [13].
⑤ The “passenger strand” is released.
⑥ The “guide strand” is integrated in the active RNA Interference Specificity Complex (RISC) that contains different argonautes and argonaute-associated proteins [16].
⑦ The siRNA guides RISC to the target mRNA.
⑧ RISC delivers the mRNA to cytoplasmic foci named processing bodies (P-bodies or GW-bodies) wherein mRNA decay factors are concentrated [17, 18].
⑨ The target mRNA is cleaved by Ago2 and degraded.
siRNA : key to sequence-specific RNA interference
Even though dsRNA were shown to induce gene-specific interference in early mouse embryos [3], preliminary attempts to use dsRNA in mammalian systems were not conclusive. These experiments employed long dsRNAs which instead of triggering RNAi generated an overall decrease in mRNA eventually leading to apoptosis, a response mediated by the interferon-activated dsRNA-dependent protein kinase [4].
Tuschl and colleagues [1] have demonstrated that this non-specific response can be bypassed by using chemically synthesized 19 to 23 nt dsRNAs [5-8]. Transfection of these siRNAs resulted in strong and sequence-specific suppression of gene expression in different mammalian cell lines.
This discovery has led to the widespread use of this technology to study mammalian gene function including clinically relevant genes, alluding to the potential therapeutic applications of RNAi-based technologies [9].
psiRNA™ plasmids, an alternative to chemically synthesized siRNAs
Chemically synthesized siRNAs are expensive and they induce only transient gene silencing due to their short life length. Another limiting step for efficient gene silencing is cell transfectability.
To overcome these limitations, InvivoGen has designed an efficient, highly transfectable and simple-to-use plasmid, called psiRNA™, that allows the production of siRNAs within the cells.
psiRNA is an RNA polymerase III-based plasmid that produces short hairpin RNAs (shRNAs). psiRNA is used to insert a DNA fragment of approximately 50 mer designed in such a way that after transcription from the human 7SK RNA polymerase III promoter it will generate shRNAs.
shRNAs are more stable than synthetic siRNAs and since they are continuously expressed within the cells, this method permits long-lasting silencing of your gene of interest.
siRNA Wizard™, free online-software to design siRNAs/shRNAs
One critical step for efficient gene silencing using RNAi is the design of siRNAs/shRNAs that greatly reduce the expression of the target transcripts without affecting unintended targets.
InvivoGen has developed a design algorithm, named siRNA Wizard, that reliably identifies siRNA/shRNA sequences for any given gene. The sequence selection is based on several criteria, such as thermodynamic stability, base composition and secondary structures.
The siRNA wizard algorithm is experimentally validated. It is used to generate ready-made psiRNAs, an expanding collection of plasmids that express shRNAs that induce silencing of their target transcript expression of >70%. The silencing efficiency of these ready-made psiRNAs is tested using the psiTEST™ system, a rapid and simple method to screen for functional siRNA or shRNA sequences.
References:
1. Elbashir S.M. et al., 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev, 15(2):188-200.
2. Fuchs U et al., 2004. Silencing of disease-related genes by small interfering RNAs. Curr. Mol. Med 4:507-517.
3. Wianny F. and M. Zernicka-Goetz, 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol, 2(2):70-5.
4. Gil J. and M. Esteban, 2000. Induction of apoptosis by the dsRNAdependent protein kinase (PKR): mechanism of action. Apoptosis, 5(2):107-14.
5. Elbashir SM. et al., 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411(6836):494-8.
6. Brummelkamp TR. et al., 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science, 296(5567):550-3.
7. Lee NS. et al., 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol, 20(5): 500-5.
8. Scherr M. et al., 2003. Gene silencing mediated by small interfering RNAs in mammalian cells. Curr Med Chem. 10(3):245-56.
9. Ryther RCC et al., 2005. SiRNA therapeutics : big potential from small RNAs. Gene Ther 12(1): 5-11.
10. Yi R. et al., 2005. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 11(2):220-6.
11. Chendrimada TP. et al., 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 436(7051):740-4.
12. Haase AD. et al., 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6(10):961-7.
13. Matranga C. et al., 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 123(4):607-2.
14. Gregory RI. et al., 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 123(4):631-40.
15. Rivas FV. et al., 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 12(4):340-9.
16. Meister G. et al., 2005. Identification of novel argonaute-associated proteins. Curr Biol. 15(23):2149-55.
17. Liu J. et al., 2005. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 7(12):1161-6.
18. Jakymiw A. et al., 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol. 7(12):1167-74.



