TLR7 and TLR8: Key players in the antiviral response
TLR7 and TLR8 are phylogenetically and structurally related. TLR7 is predominantly expressed in lung, placenta, and spleen, while TLR8 is predominantly expressed in lung and peripheral blood leukocytes, in particular monocytes.
TLR7 and TLR8 Agonists
Using TLR7-deficient mice and HEK293 cells transfected with human TLR7, Hemmi et al. have demonstrated that TLR7 is involved in the recognition of the imidazoquinoline compounds imiquimod (R837) and resiquimod (R848) [1].
Human TLR8 also mediates the recognition of R848 but surprisingly not R837. Mouse TLR8 recognizes neither of these compounds suggesting that TLR8 is nonfunctional in mice[2]. Furthermore, Lee et al. reported that guanosine analogs, such as loxoribine, activate immune cells via TLR7 but not TLR8[3].
Recently, the natural ligands of TLR7 and TLR8 were identified as single-stranded RNA (ssRNA) [4-6]. Mouse TLR7, human TLR8, and to a lesser extent human TLR7, recognize ssRNA viruses such as the influenza [5,6], Sendai [7] and Coxsackie B [8] viruses. This recognition requires the internalization of the virus and its replication to release the viral RNA into endosomes, where TLR7 and TLR8 reside. The interaction between the ssRNA and TLR7/8 triggers the recruitment of the adapter molecule MyD88 leading to the activation of NF-κB and other transcription factors and the production of proinflammatory cytokines and chemokines.
Purified viral ssRNAs or synthetic ssRNAs, complexed with cationic lipids to protect them from degradation and facilitate their internalization, can substitute for viruses. Although sequence specificity remains poorly defined, ssRNA containing poly(U)- or GU-rich sequences can stimulate TLR7 and TLR8 [4,5]. Synthetic siRNA duplexes containing a 5'-UGUGU-3' internal motif were also found to stimulate TLR7/8 [9].
According to Hornung et al., some siRNA motifs are stimulatory independently of their GU content suggesting the existence of specific sequences that are recognized by TLR7/8 [10]. Since poly(U)- or GU-rich sequences are likely to be found in non-viral RNAs, it seems that TLR7 and TLR8 are able to discriminate between self and viral RNAs by recognizing the endocytic location of viral RNAs which serves as a molecular signature. Another factor that distinguishes mammalian RNA from viral and also bacterial RNA is the level of nucleoside modification. Microbial RNA which is rarely modified is a potent activator of dendritic cells (DCs) through TLR7, TLR8, and TLR3 in contrast to mammalian RNA which is abundant in modified nucleosides [11].
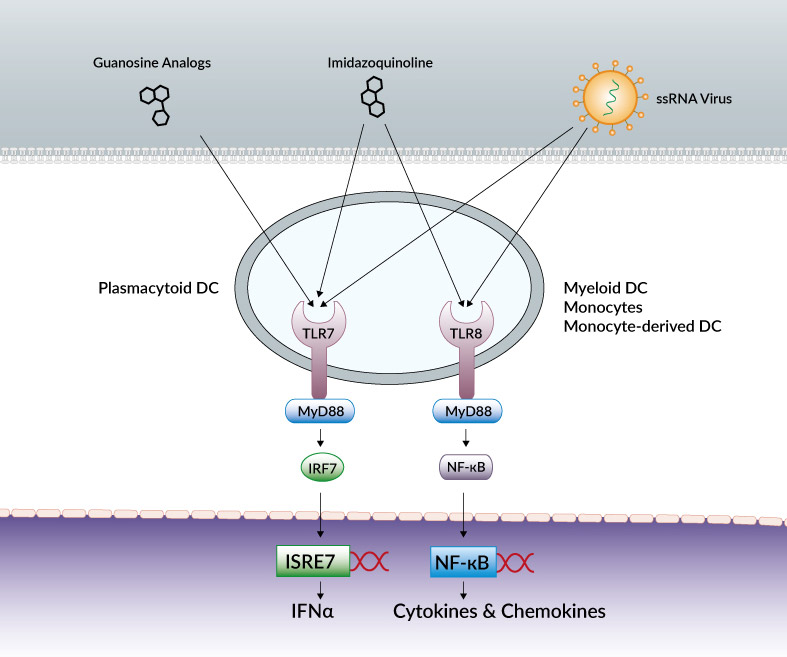
Cells activated by TLR7 and TLR8
TLR7 and TLR8 agonists differ in their target cell selectivity and cytokine induction profile. TLR7-specific agonists activate plasmacytoid DCs (pDCs) and B cells and induce mainly IFN-α and IFN-regulated cytokines.
TLR8-specific agonists activate myeloid DCs, monocytes and monocyte-derived DC leading primarily to the production of proinflammatory cytokines and chemokines, such as TNF-α, IL-12 and MIP-1α [12].
The ability of TLR7-8 agonsits to activate DCs and thus elicit Th1 and CD8+ T cells responses can be exploited to enhance the efficacy of vaccination [13,14]. They may represent better adjuvants than CpG ODN as TLR7 and TLR8 are broadly expressed on DCs and other antigen presenting cells in contrast to TLR9 which is only expressed on pDCs and B cells.
Activation of TLR7 and/or TLR8 represents a powerful and novel therapeutic strategy for the treatment of various viral infections. IFN-α is the mainstay treatment for chronic hepatitis C virus (HCV) infection but with partial success. TLR7 agonists were shown to impede anti-HCV infection by inducing IFN-α production but remain too toxic for clinical use. Thus development of new agonists is a major focus of many pharmaceutical companies.
References:
1. Hemmi H., Kaisho T. et al., 2002. Nat Immunol. 3(2):196-200.
2. Jurk M., Heil F. et al., 2002. Nat Immunol. 3(6):499.
3. Lee J., Chuang TH. et al., 2003. PNAS. 100(11):6646-51.
4. Heil F, Hemmi H. et al., 2004. Science. 303(5663):1526-9.
5. Diebold SS., Kaisho T. et al., 2004. Science. 303(5663):1529-31.
6. Lund JM., Alexopoulou L. et al., 2004. PNAS.101(15):5598-603.
7. Melchjorsen J., Jensen SB. et al., 2005. J Virol. 79(20):12944-51.
8. Triantafilou K., Orthopoulos G. et al., 2005. Cell Microbiol. 7(8):1117-26.
9. Judge AD., Sood V. et al., 2005. Nat Biotechnol. 23(4):457-62.
10. Hornung V., Guenthner-Biller M. et al., 2005. Nat Med. 11(3):263-70.
11. Kariko K, Buckstein M. et al., 2005. Immunity.23(2):165-75.
12. Gorden KB., Gorski KS. et al., 2005. J Immunol. 174(3):1259-68.
13. Craft N, Bruhn KW. et al., 2005. J Immunol.175(3):1983-90.14.
14. Wille-Reece U, Flynn BJ. et al., 2005. PNAS 102(42):15190-4.



