NOD Genes
Choose from InvivoGen's native or engineered NOD genes
InvivoGen offers a small collection of genes related to the NOD pathways. These genes can be provided in their native form, or modified by adding tags or creating dominant negative variants to facilitate diverse experimental applications.
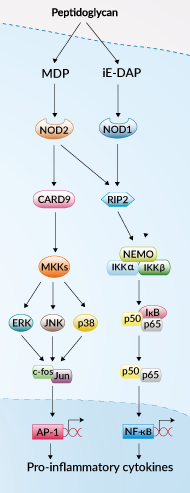
The Nucleotide-binding Oligomerization Domain-containing proteins NOD1 and NOD2 (also known as NLRs, CATERPILLERs, or NALP/PAN/PYPAFs) are cytosolic pattern recognition receptors, specialized to sense distinct motifs of peptidoglycan (PGN), an essential constituent of the bacterial cell wall. NOD1 senses the D-γ-glutamyl-meso-DAP dipeptide (iE-DAP), which is found in the PGN of all Gram-negative and certain Gram-positive bacteria. NOD2 recognizes the muramyl dipeptide (MDP) structure found in almost all bacteria, acting as a general sensor of bacterial invasion.
Key features of InvivoGen's NOD genes
- Fully sequenced genes from human and/or mouse
- Selectable in both bacterial and mammalian cells
- Simple to subclone due to unique restriction sites
- Choice of native or engineered genes (tagged, dominant negative version)
During the last few years, NOD1 and NOD2 have attracted significant attention from scientists. Genetic mutations in NOD2 were recently associated with Crohn’s disease, a chronic inflammatory bowel disease. Moreover, numerous studies have lately revealed that NOD1 and NOD2 have a close relationship with a variety of cancers via controlling proliferation, altering immunosurveillance, and interacting with tissue bacteria, including intestinal commensal intestinal microflora. Moreover, additional research into the mechanisms of NOD1 and NOD2 in cancers would shed light on the innate immunity-cancer relationship and provide intriguing targets for immunotherapy.
➔ Didn't find your gene of interest? Click here to explore InvivoGen's growing collection of over 1,800 full-length sequenced genes from A - Z.