Omicron Variants (BA.4/BA.5) Spike Pseudotyping Vector
| Product | Unit size | Cat. code | Docs. | Qty. | Price | |
|---|---|---|---|---|---|---|
|
pLV-SpikeV13 Spike pseudotyping plasmid - Omicron BA.4/BA.5 Variants |
Show product |
20 µg |
plv-spike-v13
|
|
For lentiviral particle pseudotyping with the SARS-CoV-2 Spike (BA.4/BA.5 - South Africa, Jan-March 2022)
pLV-SpikeV13 has been specifically designed for the pseudotyping of lentiviral particles with the SARS-CoV-2 Spike (S) protein when co-transfected with plasmids encoding a reporter and accessory proteins (not provided). pLV-SpikeV13 encodes the full-length Spike sequence from the Omicron variants (BA.4 and BA.5 lineages), and for optimal expression, it is codon-optimized and the C‑terminal ER-retention signal has been removed [1, 2].
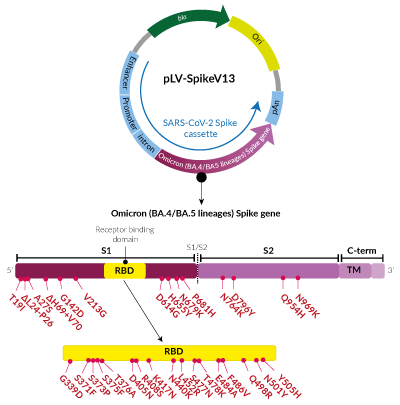
Omicron Variants (BA.4/BA.5 sublineage) Spike pseudotyping vector
 InvivoGen also offers:
InvivoGen also offers:
• SARS-CoV-2-Cellular Receptor Genes
• HEK293 ACE2-expressing Cell Line
• A549 ACE2-TMPRSS2-expressing Cell Line
Gene Description
This plasmid encodes the Spike protein from the SARS-CoV-2 Omicron BA.4 and BA.5 variants, first reported in South Africa in early 2022. These variants share the same Spike protein. They are classified as members of Clade 22A/22B (Nextstrain classification) and BA.4/BA.5 lineages (Pango classification). They are characterized by the presence of several mutations within the Spike coding region, of which, several are of concern [3,4].
- S1 domain: T19I, deletion (Δ)L24-P26, A27S, ΔH69+V70, G142D, V213G, D614G, H655Y, N679K, P681H
- RBD: G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H
- S2 domain: N764K, D796Y, Q954H, N969K
![]() Learn more about SARS-CoV-2 variants
Learn more about SARS-CoV-2 variants
General Plasmid Description
This plasmid features a potent mammalian expression cassette composed of the ubiquitous human-(h)CMV composite promoter, a rabbit β-globin intron directly upstream of the spike gene, and a rabbit β-globin polyadenylation (pAn) signal. The spike coding sequence includes the SARS-CoV-2 signal sequence and the S1/S2 furin cleavage site [5]. These plasmids are selectable with ampicillin in E. coli.
Applications
- Generation of Spike pseudotyped lentiviral particles upon co-transfection with reporter and accessory protein plasmids (not provided).
- Screening of SARS-CoV-2 inhibitors including small molecules, monoclonal antibodies, or convalescent plasma.
References:
1. Johnson, M.C. et al. 2020. Optimized Pseudotyping Conditions for the SARS-COV-2 Spike Glycoprotein. J Virol 94.
2. Ou, X. et al. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11, 1620.
3. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
4. https://outbreak.info/situation-reports
5. Coutard, B. et al. 2020. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176, 104742.
Specifications
- Origin: Omicron variants (BA.4/BA.5 lineage)
- Sequence Reference: EPI_ISL_11542604
- Codon Optimized
- ORF size: 3750 bp
-
Sequencing primers:
- Forward rbt β-globin intron: TGGTTACAATGATATACACTG
- Reverse rbt β-globin pAn: CTCAAGGGGCTTCATGATGTC -
Quality Control:
- Plasmid construct is confirmed by restriction analysis and full‑length open reading frame (ORF) sequencing.
- After purification by ion-exchange
Contents
pLV-SpikeV13 is provided as follows:
- 20 μg of lyophilized DNA
![]() The product is shipped at room temperature.
The product is shipped at room temperature.
![]() Lyophilized DNA should be stored at -20 °C.
Lyophilized DNA should be stored at -20 °C.
![]() Resuspended DNA should be stored at -20 °C and is stable for up to 1 year.
Resuspended DNA should be stored at -20 °C and is stable for up to 1 year.
Details
Spike-pseudotyping of lentiviral particles
InvivoGen's pLV-Spike plasmid collection has been designed for pseudotyping lentiviral particles with the SARS-CoV-2 Spike (S) protein. The S protein a structural glycoprotein expressed on the surface of SARS‑CoV-2. It mediates membrane fusion and viral entry into target cells upon binding to the host receptor ACE2 and its cleavage by cellular proteases such as TMPRSS2 [1]. Notably, the C‑terminal cytoplasmic tail of the S protein encodes a presumptive endoplasmic reticulum (ER)‑retention motif (KxHxx), which has previously been shown to enable the accumulation of SARS‑CoV S proteins at the ER‑Golgi intermediate compartment (ERGIC) and facilitate their incorporation into new virions [2]. The removal of this motif (d19) has been shown to increase the expression of the spike protein in pseudovirions [3,4].
Pseudotyped particle production involves the co-transfection of 293T cells with a reporter protein vector (e.g. GFP), one or several plasmids encoding the necessary lentiviral proteins, and the pseudotyping pLV-Spike plasmids. The transfected cells produce SARS‑CoV‑2 Spike (S)‑pseudotyped lentiviral particles, which can then be used to infect permissive cells, such as ACE2‑expressing HEK293-derived cells and ACE2‑TMPRSS2‑expressing A549-derived cells.
References
1. Hoffmann M. et al. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:1-16.
2. Ujike, M. et al. 2016. The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles. J Gen Virol 97, 1853-1864.
3. Johnson, M.C. et al. 2020. Optimized pseudotyping conditions for the SARS-COV2 Spike glycoprotein. J Virol. 94(21):e01062-20
4. Ou, X. et al. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11, 1620.




