COVID-19-Related Cell Lines
Designed for studying SARS-CoV-2 & developing therapeutics
InvivoGen has developed families of cell lines, derived from the human embryonic kidney 293 (HEK-293) cell line or the human A549 lung carcinoma cell line, to aid in your COVID-19 related research. They have been specifically designed to study SARS-CoV-2 infection, as well as for the development of novel therapeutics, targeting either the virus or the subsequent pro-inflammatory immune response.
Spike-expressing cell lines
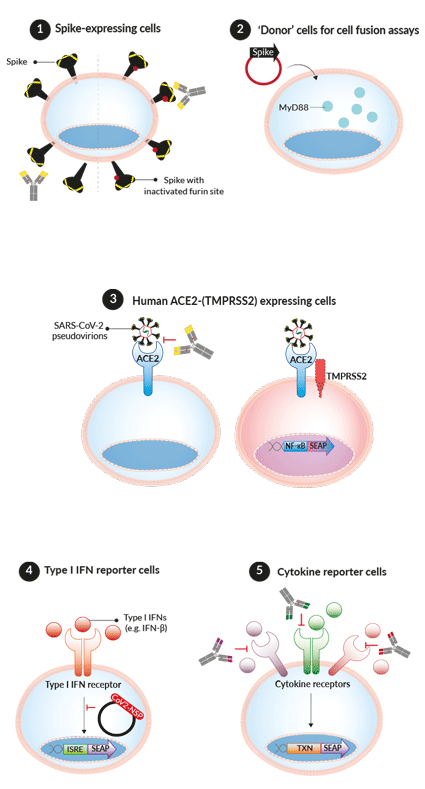
InvivoGen's various families of COVID-19-related cell lines
These cells express the SARS-CoV-2 Spike protein (Wuhan-Hu-1) either with a functional or inactivated furin cleavage site (dfur). They have been designed to screen patient/vaccinated sera and/or mAbs against the Spike by cell fusion or flow cytometry assays, respectively. Additionally, InvivoGen offers a collection of Spike expression plasmids, designed for transient or stable transfection into host cells (e.g. 293XL/null) for flow cytometry-based screening assays.
‘Donor’ cells for fusion assays
These cells have been specifically designed to study SARS-CoV-2-host cell fusion. Upon transient or stable transfection, with a Spike expression plasmid, these cells become a SARS-CoV-2 mimic, which then can fuse with InvivoGen's permissive reporter cells (below). The simple assay relies on the transfer of the adaptor molecule, MyD88, from the 'donor cell line' to the 'acceptor cell line' expressing an NF-κB-SEAP-inducible reporter gene.
SARS-CoV-2 permissive cell lines
- HEK-Blue™ hACE2(-TMPRSS2) Cells
- A549-hACE2-(TMPRSS2) Cells
- A549-Dual™ (KO-MDA5 or KO-RIG-I) hACE2-TMPRSS2 Cells
These cells express high levels of the host receptor ACE2 alone, or in combination with the host protease TMPRSS2, at their cell surface. Thus, these cells are permissive to infection by SARS-CoV-2 and Spike-pseudotyped lentiviral particles [1-3]. Of note, the addition of TMPRSS2 in A549 cells only significantly increases their infectivity [in-house data]. This cell line collection is ideal for screening small molecule inhibitors and neutralizing antibodies that aim to block the virus-host interaction and cellular signaling outcomes [4-7].
HEK cytokine reporter cells
- HEK-Blue™ IFN-α/β cells - respond to human type I interferons IFN-α & IFN-β
- HEK-Blue™ IL-6 Cells - respond to human IL-6
- HEK-Blue™ IL-1β Cells - respond to human & murine IL-1β
- HEK-Blue™ TNFα Cells - respond to human TNF-α
These cells feature a fully functional signaling pathway that is specific for human type I interferons (i.e. IFN-α and IFN-β), IL‑6, IL‑1β, or TNF‑α. They can be used to investigate how various SARS-CoV-2 proteins, such as the non‑structural proteins (NSPs), interfere with the anti‑viral interferon (IFN) response [8], as well as find ways to block the signaling pathways induced by these cytokines, which are associated with the hyper-inflammatory response triggered by SARS-CoV-2 in the more severe cases of infection [9].
![]() Read our reviews on COVID-19:
Read our reviews on COVID-19:
➤ The infection cycle of SARS-CoV-2
➤ Treatment with repurposed drugs
➤ Predicted host immune responses to SARS-CoV-2
➤ Vaccine development
➤ Protective immunity & Re-infection
References
1. Crawford, K.H.D. et al. 2020. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 12 (5):513
2. Daniloski, Z. et al. 2020. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. bioRxiv. doi:10.1101/2020.06.14.151357
3. Hoffmann M. et al. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181:1-16.
4. Ying X. et al., 2021. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Reports. 34:108628.
5. Rebendenne A. et al., 2021. SARS-CoV-2 triggers MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J. Virol. doi:10.1128/JVI.02415-20.
6. Wu J, et al., 2021. SARS-CoV-2 ORF9b inhibits RIG-I MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Reports. 34(7):108761.
7. Hu, J. et al. 2020. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Dis. doi:10.1016/j.gendis.2020.07.006
8. Banerjee, A.K. et al. 2020. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell. doi:10.1016/j.cell.2020.10.004
9. Del Valle, D.M. et al. 2020. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 26, 1636-1643.




