THP1-Dual™ KI-hSTING Cell Lines
| Product | Unit size | Cat. code | Docs. | Qty. | Price | |
|---|---|---|---|---|---|---|
|
THP1-Dual™ KI-hSTING-R232 Cells Human STING-R232 knockin - THP-1 reporter monocytes |
Show product |
3-7 x 10e6 cells |
thpd-r232
|
|
||
|
THP1-Dual™ KI-hSTING-R232 vial Additional cell vial |
Show product |
3-7 x 10e6 cells |
thpd-r232-av
|
|||
|
THP1-Dual™ KI-hSTING-H232 Cells Human STING-H232 knockin - THP-1 reporter monocytes |
Show product |
3-7 x 10e6 cells |
thpd-h232
|
|
||
|
THP1-Dual™ KI-hSTING-H232 vial Additional cell vial |
Show product |
3-7 x 10e6 cells |
thpd-h232-av
|
|||
|
THP1-Dual™ KI-hSTING-M155 Cells Human STING-M155 knockin - THP-1 reporter monocytes |
Show product |
3-7 x 10e6 cells |
thpd-m155
|
|
||
|
THP1-Dual™ KI-hSTING-M155 vial Additional cell vial |
Show product |
3-7 x 10e6 cells |
thpd-m155-av
|
|||
|
THP1-Dual™ KI-hSTING-S154 Cells Human STING-S154 knockin - THP-1 reporter monocytes |
Show product |
3-7 x 10e6 cells |
thpd-s154
|
|
||
|
THP1-Dual™ KI-hSTING-S154 vial Additional cell vial |
Show product |
3-7 x 10e6 cells |
thpd-s154-av
|
|||
|
THP1-Dual™ KI-hSTING-A162 Cells Human STING-A162 knockin - THP-1 reporter monocytes |
Show product |
3-7 x 10e6 cells |
thpd-a162
|
|
||
|
THP1-Dual™ KI-hSTING-A162 vial Additional cell vial |
Show product |
3-7 x 10e6 cells |
thpd-a162-av
|
Notification: Reference #thpd-r232-av can only be ordered together with reference #thpd-r232.
Reference #thpd-h232-av can only be ordered together with reference #thpd-h232.
Reference #thpd-m155-av can only be ordered together with reference #thpd-m155.
Reference #thpd-s154-av can only be ordered together with reference #thpd-s154.
Reference #thpd-a162-av can only be ordered together with reference #thpd-a162.
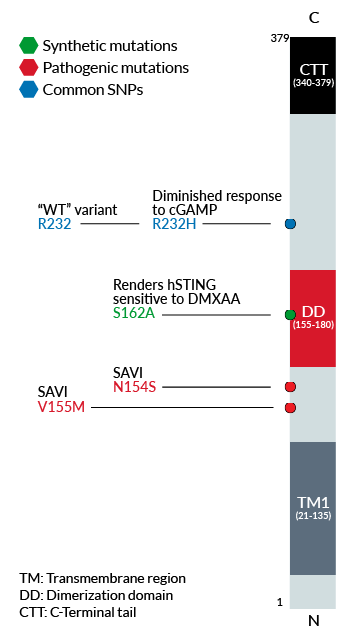
Schematic of human STING genetic variants and
their functional effect or disease association
STING knockin Dual Reporter Monocytes
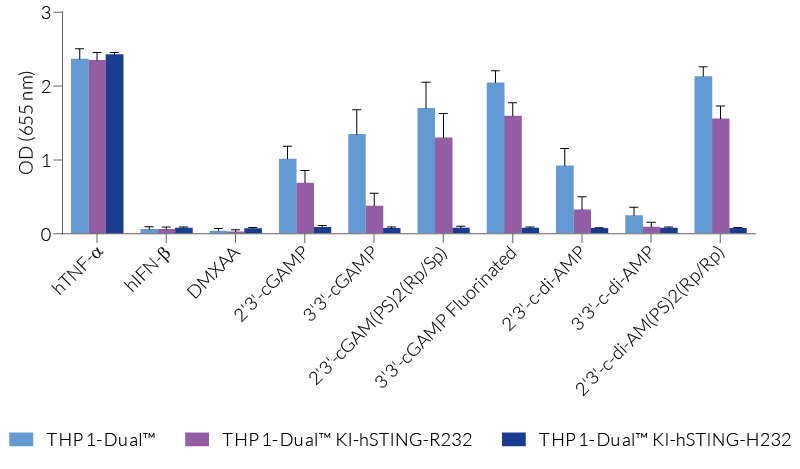
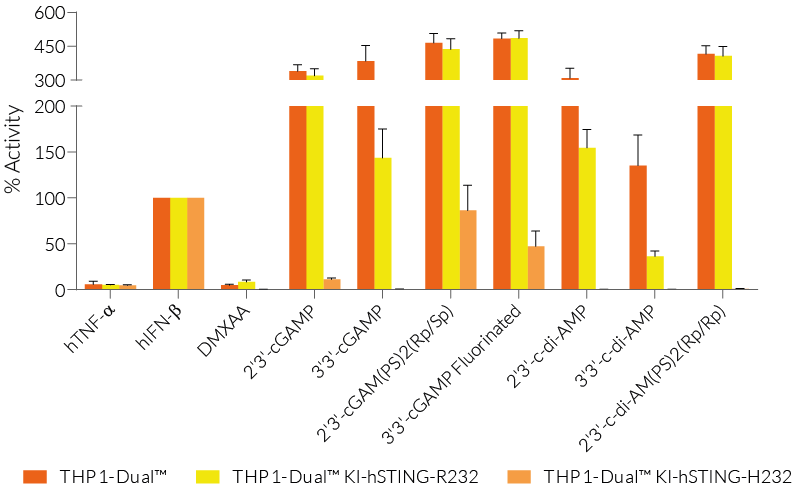
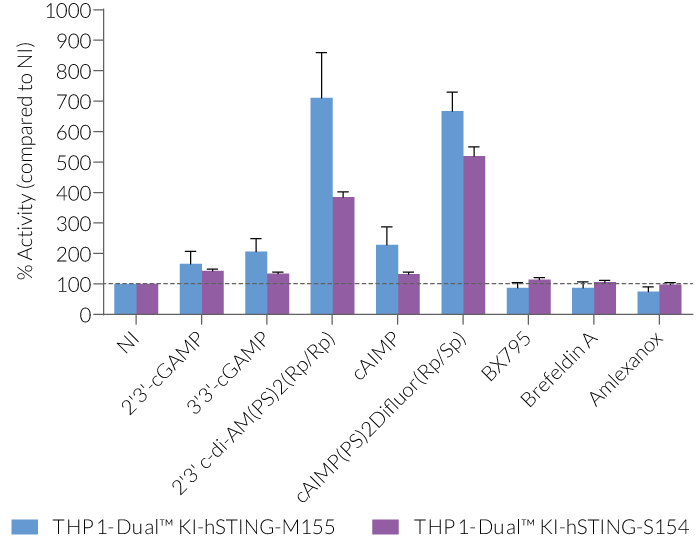
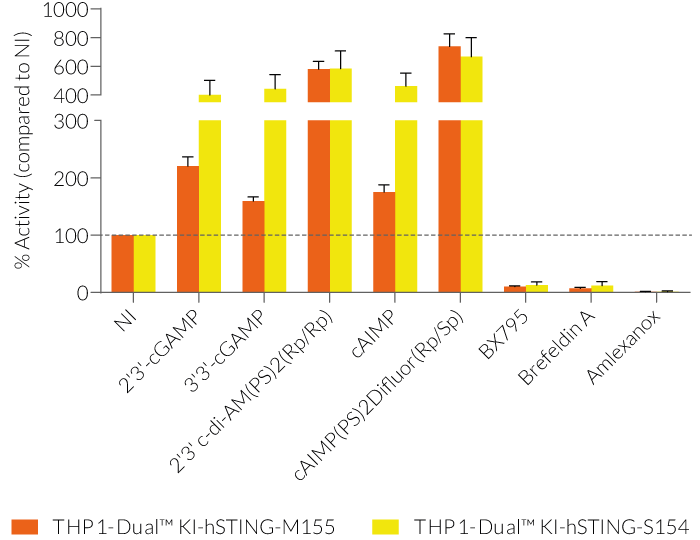
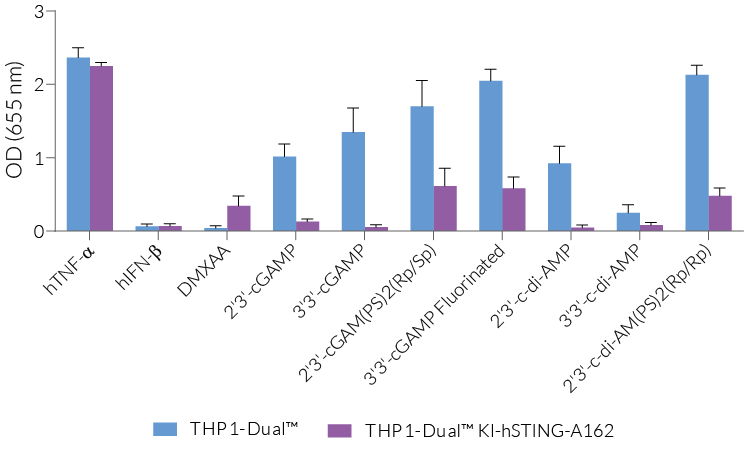
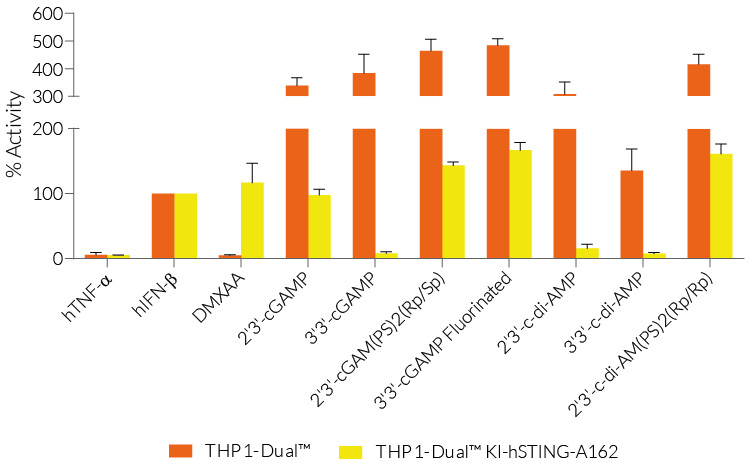
THP1-Dual™ KI-STING cells belong to a collection of human monocytic reporter cells designed to study the effect of important STING variations in the cytosolic DNA sensing (CDS) pathway.
This reporter cell family was generated from the THP1-Dual™ KO-STING cell line. They harbor a biallelic knockout (KO) of the endogenous HAQ hSTING gene, which contains three single nucleotide substitutions (SNS): H71‑A230-R232-Q293. Additionally, they express two inducible reporter genes, allowing the simultaneous study of the NF-κB and IRF pathways, by monitoring the respective SEAP (secreted embryonic alkaline phosphatase) and Lucia luciferase activities in the cell supernatant.
Each cell line features a stable knock-in (KI) of one distinct human (h)STING variant:
-
R232 – variant (R71‑G230-R232-R293) is present in ~60% of the human population and therefore, considered the wild-type (WT) hSTING [2].
-
R232H – frequently occurring isoform in the human population (~14%) leading to a diminished response to CDNs (cyclic dinucleotides) [2].
-
V155M – gain-of-function (GOF) mutation causing constitutive STING activation and severe SAVI disease (STING-Associated Vasculopathy with onset in Infancy) [3].
-
N154S – GOF mutation causing constitutive STING activation and severe SAVI disease [3].
- S162A – point mutation that renders hSTING highly sensitive to DMXAA, a potent tumor vascular disrupting agent in mice [1].
These gene variants (alleles and mutants) differ in their responsiveness to microbial CDNs leading to loss or gain of STING functionality (see figures and VDS). Therefore, they represent a powerful tool to further understand STING biology and its impact on human health [1-3].
Key features:
- Verified KO of the endogenous hSTING gene
- Stable KI of one particular hSTING variant
- Distinct monitoring of NF-κB or IRF activation by assessing the SEAP and Lucia luciferase activities
Applications:
- Defining the responsiveness of each hSTING variant to microbial CDNs
- Screening for hSTING-variant specific agonists and antagonists (H-151)
- Highlighting the impact of each genetic variation on the STING-dependent IRF and NF-kB responses.
References:
1. Gao P. et al., 2014. Binding-pocket and lid-region substitutions render human STING sensitive to the species-specific drug DMXAA. Cell Reports. 8:1668-76.
2. Yi G. et al., 2013. Single nucleotide polymorphisms of human STING can affect the innate immune response to cyclic dinucleotides. PLOS One. 8:e77846.
3. Liu Y. et al., 2014. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 371:507-18
Specifications
Growth medium: RPMI 1640, 2 mM L-glutamine, 25 mM HEPES, 10% (v/v) fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin, 100 µg/ml Normocin™
Antibiotic resistance: Blasticidin and Zeocin®
Quality Control:
- The knockin (KI) of the human STING variant has been verified by PCR, DNA sequencing, and functional assays.
- Reporter activity has been validated by stimulating the cells with STING ligands, such as c-di-AMP and cGAMP.
- The stability for 20 passages, following thawing, has been verified.
- These cells are guaranteed mycoplasma-free.
Contents
Please note: Each cell line is sold separately. See TDS for the exact contents of each cell line.
- 1 vial of THP1-Dual™ KI-hSTING variant (3-7 x 106 cells) in freezing medium
- 1 ml of Normocin™ (50 mg/ml). Normocin™ is a formulation of three antibiotics active against mycoplasmas, bacteria and fungi.
- 1 ml of Zeocin® (100 mg/ml)
- 1 ml of Blasticidin (10 mg/ml)
- 1 tube of QUANTI-Luc™ 4 Reagent, a Lucia luciferase detection reagent (sufficient to prepare 25 ml)
- 1 ml of QB reagent and 1 ml of QB buffer (sufficient to prepare 100 ml of QUANTI-Blue™ Solution, a SEAP detection reagent)
![]() Shipped on dry ice (Europe, USA, Canada, and some areas in Asia)
Shipped on dry ice (Europe, USA, Canada, and some areas in Asia)
Details
STING
The STimulator of INterferon Genes STING, alternatively known as MPYS, TMEM173, MITA, and ERIS, is a key sensor of cytosolic nucleic acids. Initially thought to serve solely as an adaptor protein for mediating signaling by cytosolic DNA sensors (CDS), STING was recently found to be a direct sensor of cyclic dinucleotides (CDNs) [1].
STING signaling
CDNs are ubiquitous second messenger molecules in bacterial or metazoal signal transduction and are defense triggers in mammalian cells. Namely, cyclic diguanylic acid (c-di-GMP), cyclic diadenylic acid (c-di-AMP), and cyclic adenylicguanylic acid (cGAMP) are the most prevalent intracellular signaling intermediates in bacteria and/or metazoa [2-3].
A direct interaction between DNA and STING could not be demonstrated, suggesting the intervention of at least one additional protein [1]. The identity of the major dsDNA cytosolic sensor was resolved in 2013: cGAs (cyclic GMP-AMP synthase) is activated upon direct DNA binding and subsequently catalyzes the production of a non-canonical cGAMP, which in turn, activates STING [3].
Once activated, STING and TANK-binding-kinase-I (TBK1) interact to induce an active interferon regulatory factor (IRF3) dimer which then binds to interferon-stimulated responsive elements (ISRE) in the nucleus and leads to IFN-α/β production [4]. The production of NF-κB-dependent inflammatory cytokines is also observed downstream of STING activation but the underlying mechanisms remain opaque [5].
STING variants and pathologies
Interestingly, a variety of natural variants of human STING (hSTING) have been identified. These gene variants differ in their responsiveness to microbial CDNs leading to the loss or gain of STING functionality. For instance, the variant R232H is drastically less sensitive to CDNs than the most prevalent STING variant 232R (~60% of the human population) [6]. In contrast, several GOF mutations (e.g. N154S and V155M) in exon 5 lead to constitutive STING activation causing severe STING-Associated Vasculopathy with onset in Infancy (SAVI). STING is also implicated in many other health disorders such as infectious diseases, cancer, and autoimmunity, and is therefore considered a promising therapeutic target [7].
References:
1. Burdette DL. et al., 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478(7370):515-8.
2. Woodward JJ. et al., 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328(5986):1703-5.
3. Wu J. et al., 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science, 339(6121):826-30.
4. Ishikawa H. et al., 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788-92.
5. Abe T. and Barber G.N., 2014. Cytosolic-DNA-Mediated, STING-Dependent Proinflammatory Gene Induction Necessitates Canonical NF-κB Activation through TBK1. Journal of Virology 88:5328-41
6. Yi G. et al., 2013. Single nucleotide polymorphisms of human STING can affect innate immune response to cyclic dinucleotides. PLOS One. 8:e77846.
7. Liu Y. et al., 2014. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 371:507-18.